
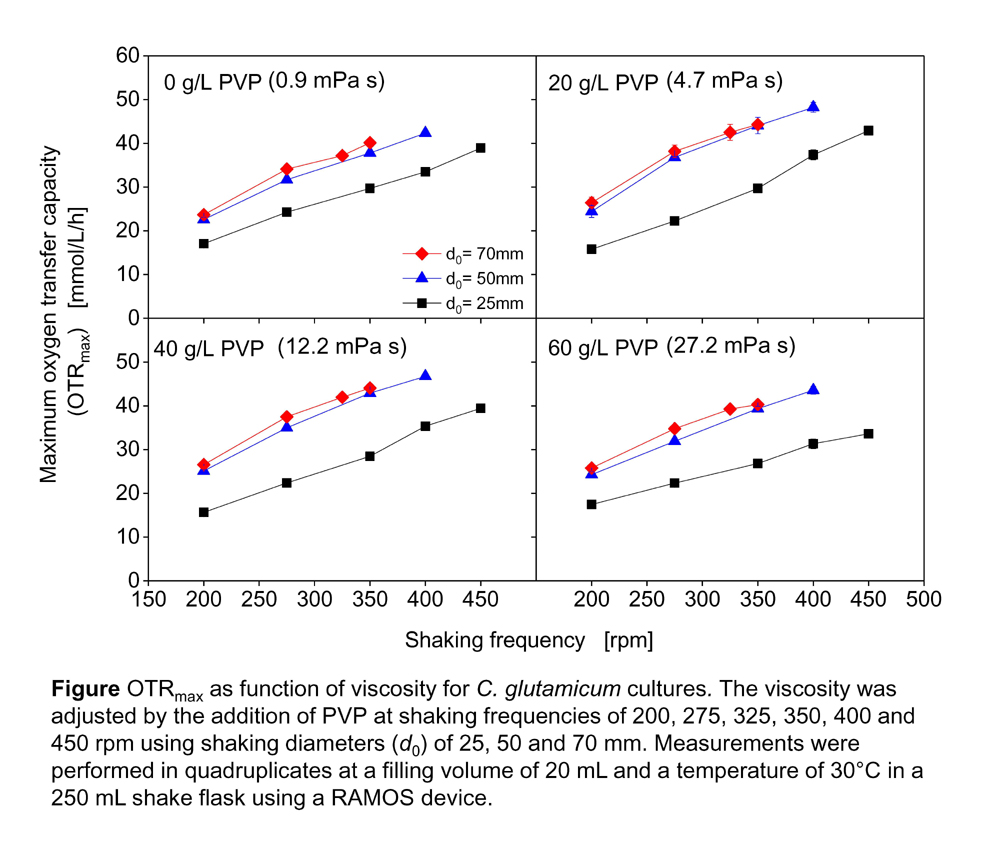
Oxygen supply is a crucial parameter in biotechnological production. While oxygen transfer has been well studied for viscous cultivations in stirred tanks, little is known about the oxygen transfer in shake flask cultures with elevated viscosity media. In our previous study, it was experimentally shown that the maximum oxygen transfer capacity OTRmax in shake flasks increased with increasing viscosities up to 10 mPa s. It was further demonstrated that the increased thickness of the liquid film at the shake flask wall inherently promotes the oxygen supply to the medium at moderately elevated viscosities1). Although the shaking diameter is an important parameter to determine the oxygen transfer in shake flask culture, is not well discussed in the past.
In this study, OTRmax was investigated using a biological model system. Corynebacterium glutamicum DM1730 was studied in culture medium with different shaking diameter (25, 50, 70 mm) at elevated viscosity (1-27 mPa s), using polyvinylpyrrolidone as viscosifying agent. The oxygen transfer was monitored using a respiration activity monitoring system (RAMOS)2) with 20 mL medium in 250 mL flask. As a result, the OTRmax significantly increased by changing shaking diameter from 25 to 50 mm under each viscosity condition. It was hypothesized that with the increase of shaking diameter, a larger part of the shake flask wall became available for the liquid film and the bulk liquid was more pressed into one corner of the flask. Meanwhile, no further increase of OTRmax was observed by changing the shaking diameter from 50 to 70 mm. It was suggested that the volume of liquid film already reached the maximum at 50 mm shaking diameter under the examined condition.
1) Giese et al., Biotechnol. Bioeng., 111, 295-308 (2013)
2) Anderlei et al., Biochem. Eng. J., 17(3), 187-194 (2004)
Natural media are often used for various commercial bioprocesses by manufacturers to cut raw material cost. However, the components of the raw materials varies between lot-to-lots and brand-to-brands. The varieties of raw materials influence to the cell growths and materials productivities, and results in unstable production across batches in manufacturing processes. To ensure the quality of raw materials among batches, it is necessary to perform a laboratory screening to purchasing the optimal one, and ensure a desirable performance in industrial process. To solve the serious problems in bioprocesses, it is developing that a modeling methodology using composition of raw materials, named us “substratome”(1), obtained by non-targeted metabolomics-like methods can estimate the cell growth and bio-productions. Here, we will present that two model studies: [1] Escherichia coli growths have been estimated from hydrophilic components in yeast extract obtained by gas chromatography-mass spectrometry (GC-MS)(1), and [2] bioethanol production have been estimated by the volatile components in corncob and corn stover hydrolysates obtained by GC-MS; by partial least square regression (PLS-R)(2). Additionally, we will present preliminary results to solve the same issues by using artificial intelligence.
References
1) Tachibana S, Watanabe K, Konishi M. (2019) J. Biosci. Bioeng. (in press)
2) Watanabe K, Tachibana S, Konishi M. (2019) Bioresouc. Technol. 281: 260-268
Cancer is a complex disease that relies on both oncogenic mutations and non-mutated genes for survival, thereafter coined as oncogene and non-oncogene addictions. The need for more effective combination therapies to overcome drug resistance in oncology has been increasingly recognized, but the identification of potentially synergistic drugs at scale remains challenging. Here we propose a gene-expression-based approach, which uses the recurrent perturbation–transcript regulatory relationships inferred from a large compendium of chemical and genetic perturbation experiments across multiple cell lines, to engender a testable hypothesis for combination therapies. These transcript-level recurrences were distinct from known compound–protein target counterparts, reproducible in external datasets, and correlated with small-molecule sensitivity. We applied these recurrent relationships to predict synergistic drug pairs for cancer, and experimentally confirmed the synergistic effects of one combination of a retinoid and an mTOR inhibitor and the other combination of a protein synthesis inhibitor and a CDK inhibitor in two breast and two lung cancer cell lines. Our results corroborate a gene-expression-based strategy for combinatorial drug screening as a way to target non-mutated genes in complex diseases.
Recombinant proteins expressed in E. coli are frequently misfolded into insoluble aggregates called inclusion bodies, which require a complicated downstream process for purification including a refolding step. Therefore, the increase of solubility of expressed proteins has been a critical issue in recombinant protein production using E. coli. It is known that the solubility is increased by lowering the protein synthesis rate under proper DO environment. In this study, we developed a DO-stat fed-batch culture system, which allows to maintain DO in a fermenter constant by glucose feeding. Using the DO-stat fed-batch system, anti-CRP scFv production by E. coli employing the pET expression system was investigated. By employing a PID control to determine glucose feeding rate, DO in a fermenter was successfully maintained. The results of the fed-batch operation at DO=40% showed the solubility of anti-CRP scFv improved from 23-29% by flask and batch cultures to approximately 100%. The anti-CRP scFv production also increased from 0.14 g/L by a batch culture to 2.6 g/L. DO in a fermenter significantly influenced on the solubility and production of scFv and the optimal DO level was found to be between 40-60% for successful scFv production. It was also found that the increased addition of yeast extract was effective to increase anti-CRP scFv production by avoiding the proteolytic degradation of scFv that sometimes happened in the mid or later phase of fed-batch cultivation. Thus, it was found that the DO-stat fed-batch culture system greatly improves the solubility and production of anti-CRP scFv using E. coli.
The characterization of polyphenol oxidase (PPO) taken from the seed kernel of the Philippine mango (Mangifera indica var. Philippines) was studied. PPO was obtained by extraction followed by partial purification through ammonium sulphate precipitation. The partially purified PPO has an optimum pH of 7.0 and an optimum temperature of 30 °C using catechol as substrate. The PPO showed maximum activity when catechol was used as the substrate, followed by other diphenol substrates 4-methylcatechol, L-DOPA, and hydroquinone. Meanwhile, no enzyme activity was detected from two monophenol substrates p-cresol and tyrosine. In the determination of kinetic parameters using catechol as substrate, the Michaelis-Menten constant (Km) was found to be 2.50 mmol/L while the maximum reaction velocity was 0.12 U/min.