AMBIC (AMBIC.org) is an industrial–academic–government consortium dedicated to addressing research and development challenges in upstream cell culture bioprocessing identified by the 22 industrial and 2 government members. Focal areas of the center include industrially-relevant biology, process monitoring and control, and consensus and standardization issues. Proposed areas of common interest to industrial members are elucidated through summer workshops organized around the topical areas of cells, raw materials, processes, and equipment, resulting in a call for proposal to academic members. Academics then present proposals which are voted on by industrial members to yield 2-year research and development projects in academic labs that address these biomanufacturing challenges under the guidance and mentorship of our industrial sponsors during monthly meeting. These activities have resulted in the start-up and execution of approximately 18 active and several graduated research projects under way at the academic participating institutions including Johns Hopkins University, University of Delaware, University of Massachusetts, Lowell, Clemson University, and University of Maryland, College Park. This presentation will discuss the activities of the AMBIC center followed by an overview of the projects underway within AMBIC. Projects currently under way include efforts to develop a community reference cell, process, and media, elucidating the solubility of nutrient concentrates and chemical complexes, understanding and manipulating the epigenome, identification of inhibitory waste products, sensors development for Surface Enhanced Raman Spectroscopy (SERS) and “Smart Marbles”, adaptation of process models for cell culture media and process feeding, controlling glycolytic pathways through chemical additives, and evaluating extracellular vesicles, ER stress pathways, and host cell proteins,. Progress obtained on these projects will be discussed along with a description of how the biopharma ecosystem is partnering and collaborating to identify and address critical common biomanufacturing challenges.
The monoclonal antibodies (mAbs) are widely used to produce therapeutic proteins. The rapidly evolving market for mAbs including biosimilars is making it necessary to realize superior manufacturing processes regarding costs and time. Generally, production in continuous mode can improve process efficiency. Continuous manufacturing for mAbs is intensively researched for individual unit operations in the upstream (e.g., perfusion) and downstream (e.g., chromatography using simulated moving bed), and for the end-to-end process. However, it is yet to establish process design models that can indicate when the novel continuous mode becomes more beneficial than the conventional batch mode.
We present a simulation-based approach for designing mAb production processes considering the continuous mode as an alternative. Differential equation models for cell cultivation and protein A separation have been applied based on fundamental physical phenomena. The model parameters were estimated using production data from a pilot scale research facility for the production of mAbs from Chinese hamster ovary (CHO) cells. After confirming the model performance using the production data, we performed economic process assessment. Different scenarios were considered regarding the operation modes, i.e., batch or continuous, and equipment, i.e., single-use or multi-use, for the individual units as well as the end-to-end process.
The simulation results indicated that the favorable scenarios varied in the upstream and downstream depending on the production scale and the CHO cell performance. The superiority of the fully integrated continuous process has been seen regarding productivity. Sensitivity analysis revealed the relevance of the cell performance, which was represented by the maximum doubling and death rates in the model. This finding will open up a new research opportunity to connect cell design with product and process design on a simultaneous basis.
Chinese hamster ovary (CHO) cells have been widely used as a host for biopharmaceutical protein production. Usually, producer cell lines are established from a recombinant cell pool containing thousands of cells with random integration of transgene. Thus, the screening of producer cells is time-consuming and labor-intensive process. We have previously reported an accumulative transgene integration system (AGIS) using Cre-recombinase and mutated loxPs, in which transgenes can be efficiently introduced into a chromosomal locus carrying a loxP target site [Kameyama et al., Biotechnol. Bioeng., 105, 1106–114 (2010); Obayashi et al., J. Biosci. Bioeng., 113, 381–388 (2012)]. In this study, the system was applied for establishing high-producer CHO cells. The founder cells, in which a mutated loxP and an expression cassette encoding the red fluorescent protein (DsRed) were introduced into the hprt locus of CHO cell genome, were used. An expression cassette of anti-prion scFv-Fc was introduced into the donor plasmids. Cells were transfected with donor plasmids and Cre-expression vector by lipofection or electroporation. To accelerate the screening process, the target cells were selected by change in color of reporter fluorescent proteins. Simultaneous transgene integration from two donor vectors was possible for the screening of cells with multiple copies of transgene. A minicircle DNA vector as a donor facilitated the transgene integration. We obtained targeted CHO cells with 6 copies of transgene into the hprt locus after three rounds of transfection and screening. The scFv-Fc productivity increased corresponding to the transgene copy number. This procedure is promising for the generation of producer cell lines for the production of pharmaceutical recombinant proteins.
This work was supported in part by grants for developing key technologies for discovering and manufacturing pharmaceuticals used for next-generation treatment and diagnoses, both from METI and AMED under Grant Number JP17ae0101003.
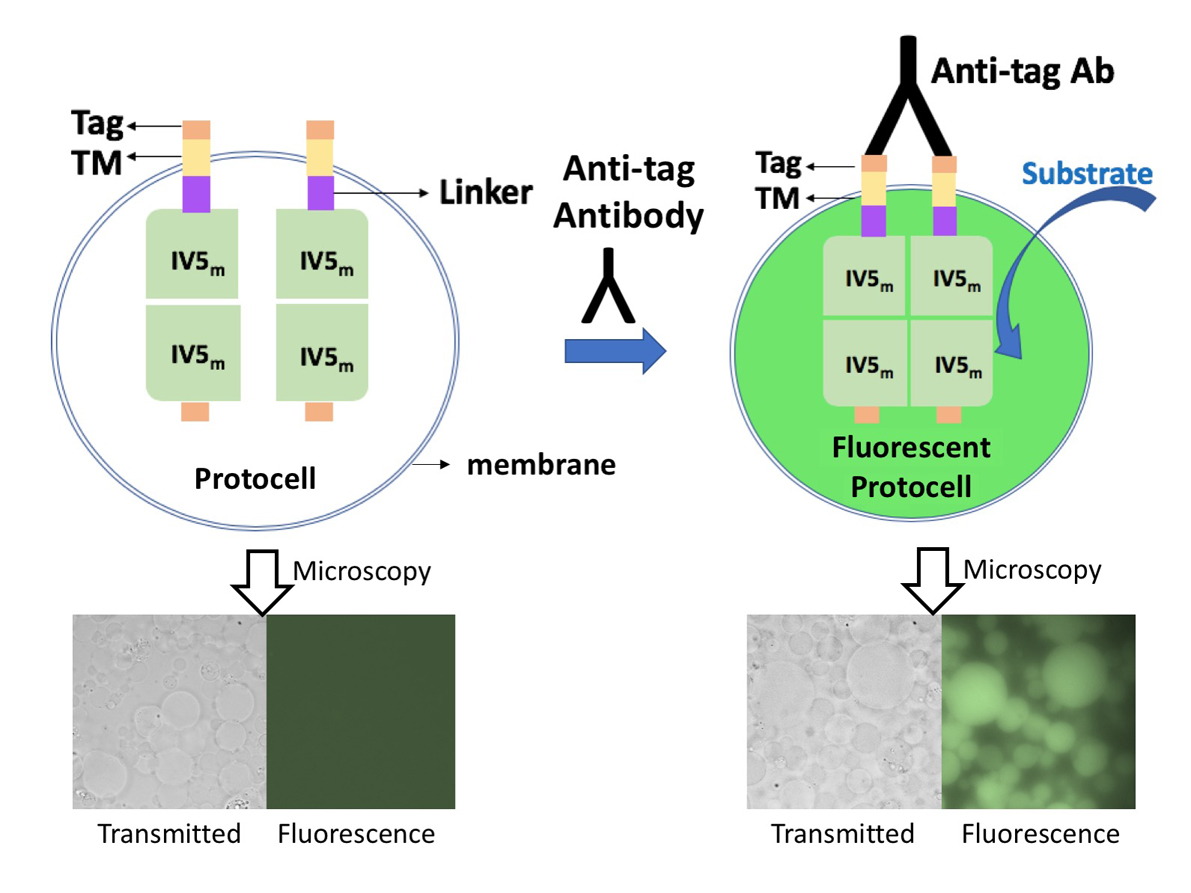
Digital counting of fluorescent signals generated in many small compartments can significantly improve the detection sensitivity of quantitative PCR and enzyme-linked immunosorbent assay (ELISA). However, the reported digital ELISA systems still need extensive washing steps to remove background signal, which hampered its performance. To address this problem, creation of a vesicle (Protocell) array that amplifies the binding signal of external protein as a fluorescent signal was attempted. We chose mutant β-glucuronidase (GUS) as a reporter enzyme that needs assembly of the four subunits by the dimerization of dimers, and which is inhibited by a set of interface mutations. Using a thermostabilized GUS mutant IV-5, we screened out an interface mutant (M516K, F517W) to create IV5m - a mutant with high thermostability and activity that is inactive on its own but becomes active when dimerized. After tethering short N-terminal tag and transmembrane (TM) sequences derived of epidermal growth factor receptor, the fusion protein was expressed by a cell-free protein synthesis system inside protocells, which were formed by the centrifugation of inverted emulsion. When corresponding tag-specific antibody was applied outside of the protocells, a clear increase in GUS activity was observed inside vesicles by adding fluorescent substrate, probably due to spontaneous integration of the expressed protein into the vesicles and dimerization by the antibody bound to the displayed tag. Furthermore, using a flow cytometry (FCM) digital signal was obtained by counting fluorescent protocells in response to external anti-His6 antibody concentration. Furthermore, SpyTag/SpyCatcher system was tested on this biosensor by expressing SpyTag-TM-IV5m in protocells, and adding His6-tagged SpyCather protein and anti-His6 antibody outside of protocells. An antibody dose-dependent increase in the population of fluorescent protocells was also detected by FCM, implying its potential applicability to antigen detection system. We are now testing the single molecule detectability of the system.
Acknowledgement: SICORP of JST.
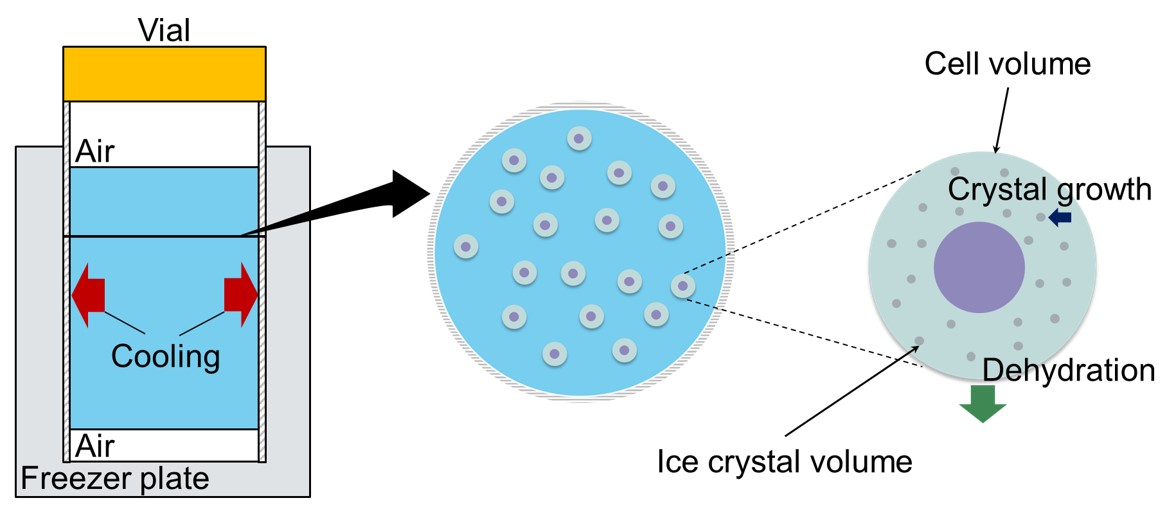
In the last years, the demand for human induced pluripotent stem (hiPS) has increased along with the progress of the clinical trials. This trend indicates the necessity for establishing freezing processes of hiPS cells for preservation and transportation. Generally, there are two methods for freezing cells: slow freezing and vitrification. In the field of cryobiology, both experimental and modeling studies have been conducted on slow freezing. The process can benefit from scalability, simplicity in operation, and no direct contact with freezing agent. However, the process could suffer from the quality variation intracontainer, and intra- and interfreezer.
We present a model-based design of slow freezing processes for hiPS cells considering the quality variation inside a container (i.e., a vial). A single-cell model was developed that integrated mechanistic models describing a radial and temporal temperature profile, cell volume change through transmembrane water transport, and intracellular ice formation during slow freezing. The overall inputs of the model are the cooling rate of the freezer, the vial diameter and material, and the cryoprotective agent type. The outputs are the maxima of cell volume change and the intracellular ice crystal volume as the cell quality indicators, and the required freezing time as the productivity indicator.
The model showed sufficient performance compared with a recently reported experimental study on hiPS cells. Based on this confirmation, the model was applied for three design cases. The choice of a cryoprotective agent, cooling rate, and vial diameter significantly affected the quality evaluation. When considering the productivity, the optimal vial diameter changed depending on the cell demand and the acceptable intracellular ice crystal volume. In the ongoing work, we are performing rigorous performance assessment of the model by extensive experiments. Advanced process optimization such as dynamic temperature optimization is another ongoing subject.