Introduction
The safe and convenient yeast Saccharomyces cerevisiae is widely studied for the production of chemicals. D-Lactic acid is used as one of the biodegradable plastic materials. D-Lactic acid production by the engineered microorganisms often requires neutralization of the culture medium and the addition of neutralization reagents, which increase the cost of D-lactic acid production considerably. Previously, we developed a method improving various tolerances such as thermotolerance of yeast through CRISPR-Cas mediated genome evolution. In this study, a lactic acid tolerant yeast was constructed by this method and the resultant yeast was metabolically engineered to obtain a yeast producing D-lactic acid.
Method
First, we constructed a mutant library of lactic acid tolerant yeast by genome evolution method and screened a lactic acid tolerant yeast. Secondly, we integrated glycolytic enzyme genes and D-ldh (from Leuconostoc mesenteroides) into genome DNA of the lactic acid tolerant yeast to modify the expression of these genes. Then, the best strain producing D-lactic acid was screened from the transformants. Finally, we cultured the resultant strain in a non-neutralizing condition, and its fermentation ability and transcription levels of glycolytic enzyme genes and D-ldh were investigated.
Result
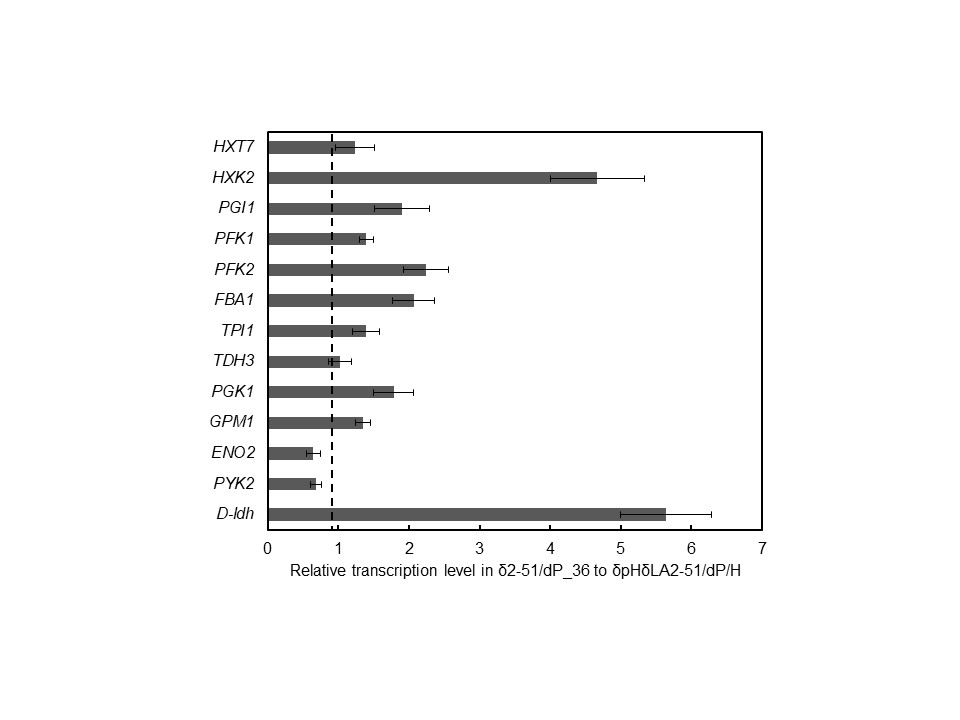
We selected δpHδLA2-51 as a lactic acid tolerant yeast, which could grow on YPD+60 g/L lactic acid medium. Engineered strain δ2-51/dP_36, which was constructed by modifying the expression of the glycolytic enzyme genes and D-ldh in δpHδLA2-51, produced D-lactic acid in a yield of 0.30 g/g-glucose in non-neutralizing condition. In the log phase, δ2-51/dP_36 showed higher transcription levels of 9 glycolytic enzyme genes and D-ldh compare to control strain δpHδLA2-51/dP/H.
Acknowledgements
This work was partly supported by JSPS KAKENHI (grant number JP18K14069 and JP18KK0413).
Biodiesel fuel is monoalkyl esters of long-chain fatty acids, which is synthesized by bio-based oils and alcohols as alternatives for fossil fuels. Biodiesel fuel production is usually catalyzed by an alkali catalyst. The use of an enzymatic catalyst allows the use low-grade oil feedstock containing a high percentage of free fatty acids, easy recovery of glycerol, mild reaction conditions, and reduced wastewater. However, an obstacle of enzymatic methanolysis is the limited stability of lipases in the presence of methanol. Therefore, in this study, we improved the stability of the lipase from Thermomyces lanuginosus DSM 10635 in methanol by using protein engineering methods1). The plasmid pE_TLLHis6N, which was constructed by cloning the lipase from T. lanuginosus DSM 10635, was used for producing the wild-type lipase and as a template for constructing site-specific mutant lipases. Escherichia coli BL21 (DE3) cells transformed with the pE_TLLHis6N plasmid (or its derivatives) were cultured in LB liquid media containing 50 mg/L ampicillin sodium salt. The protein expression was induced by the addition of isopropyl-β-D-1-thiogalactopyranoside (IPTG). The proteins were purified by using HisTrap column. The purified enzyme solutions were incubated in the presence of 50% (v/v) methanol at 40°C and the remaining activity was measured. Figure shows the time courses of the remaining activity of the wild-type and mutated lipases in the presence of methanol. Deactivations of the wild-type and mutated lipases obeyed first order kinetics for 0.5~2.0 h. The half-lives of three mutated lipases (A28S, A163D and F174Y) were longer than those of the wild-type lipase. In particular, the half-lives of the A28S were observed to be 6.5 times longer than those of the wild-type lipase. The half-lives of three mutated lipases (A28S, A163D and F174Y) were successfully improved by using protein engineering methods.

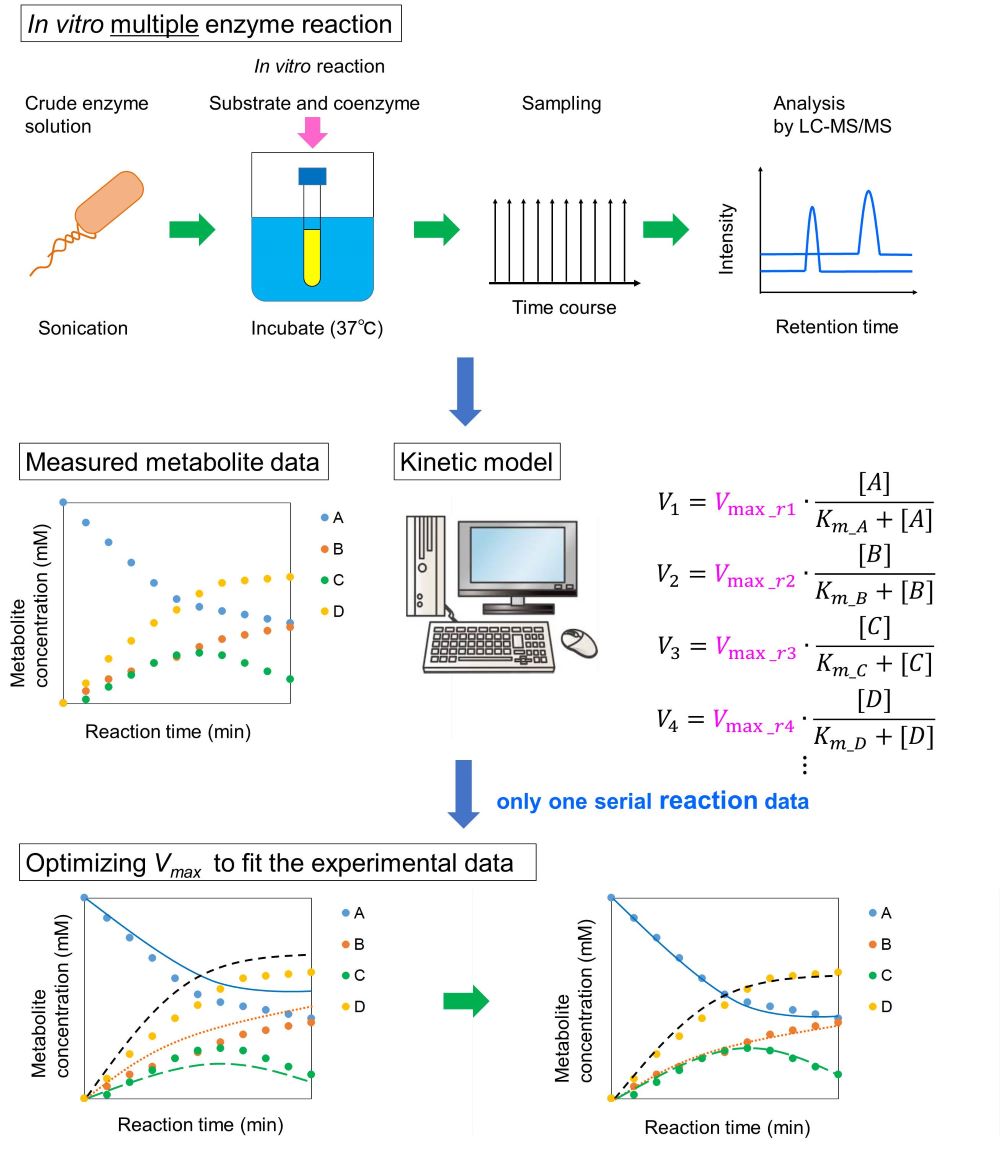
Many useful compounds such as, fuel alcohols, plastic and polymer materials, have been produced via the metabolism in microbial cell factories. In order to improve the conversion rate from substrate to the desirable target, it is required to identify and eliminate a rate limiting reaction in the pathway. Specific enzyme activity (Vmax), the parameter representing the maximum reaction rate, is one of the important clues for identifying the rate-limiting reaction. By comparing the Vmax value with actual metabolic flux for each reaction, it should be possible to identify a rate-limiting reaction in the pathway. However, because conventional method for measuring Vmax requires different assay protocols for each enzyme, it was time consuming to investigate multiple enzymes. In this study, we developed a method to simultaneously measure the Vmax values of multiple enzymes in a pathway using a kinetic model with a time course of the intermediate metabolite concentration in vitro experiment. To test the proof of concept, we estimated Vmax values for 9 glycolytic enzymes in Escherichia coli using a previously reported kinetic model and a hypothetical time course data which was generated from a simulation using the model with an arbitrary Vmax set. The parameters of Vmax were optimized to fit the experimental time-series data of the intermediate metabolites. As a result, we successfully estimated the initially set Vmax values. Here we demonstrated the Vmax values estimation using a crude cell extract obtained from wild type E. coli cells. In vitro reaction experiment was performed using the crude cell extract with addition of glucose and essential cofactors. After starting reaction, a sample was collected at every 10 minute and was analyzed with a liquid chromatography tandem mass spectrometry to obtain the time series of intermediate metabolites. In the poster presentation, we would like to discuss more specific research contents.
para-Aminobenzoic acid (PABA) is a building block for pharmaceuticals, and it has a great potential to serve as a raw material for aromatic polymers including engineering plastics. Currently, it is produced mainly by multi-step chemical conversions from petroleum-derived toluene via p-nitrotoluene and p-nitrobenzoate. With the recent increases in environmental consciousness, there is a growing interest in the production of PABA from renewable resources by fermentation. Escherichia coli can synthesize PABA from glucose via the central metabolic pathway shikimate pathway and folate pathway. Therefore, in this study, we aimed PABA production from glucose with E.coli. Firstly, we introduced into E.coli two genes pabAB and pabC which express aminodeoxychorismate synthase and 4-amino-4-deoxychorismate lyase respectively and used this engineered strain to produce PABA from glucose. The strain produced up to 1.92 g/L of PABA from 20 g/L glucose. Secondly, to increase PABA production, we tried to enhance the shikimate pathway of this engineered strain by introducing aroA, aroB, aroC, aroD, and pA1lac promoter-controlled aroFfbr respectively. However, these strains exhibited no increase in PABA production. Thirdly, we tried to enhance the folate pathway of E. coli overexpressing pabAB and pabC genes by adding Glutamine at 10 mM. As a result, by adding Glutamine at 10 mM, PABA production increased up to 3.03 g/L. This result suggests that the folate pathway is the rate-limiting step and the folate pathway needs to be enhanced to further increase PABA production.
Introduction
Although, yeast Saccharomyces cerevisiae has been used to produce various bio-based chemicals including solvents and organic acids, most products inhibit their growth at high concentrations. In general, it is difficult to rationally improve stress tolerance in yeast by modifying specific genes because many unspecified genes are involved in stress response. Previous studies reported that various stress tolerance in yeast was improved by introducing random mutations such as DNA point mutations1) and DNA structural mutations. In DNA point mutagenesis, protein functions and activities can be altered by changing several bases. On the other hand, in DNA structural mutagenesis, expression levels of multiple proteins can be altered by changing a wide range of bases. In this study, we developed a novel mutagenesis strategy combining point mutagenesis and structural mutagenesis to construct yeast with high 2,3-butandiol (2,3-BDO) tolerance.
Results
A DNA point mutation and a structural mutation were simultaneously introduced into S. cerevisiae YPH499 strain while stepwise increasing the concentration of 2,3-BDO. Resultant mutant YPH499/pol3δ/BD_392 showed 1.3- and 2.6-fold higher cell concentrations than parent strain YPH499 after 96 h cultivation in media containing 125 and 150 g/L of 2,3-BDO, respectively.
Conclusion
We successfully developed the novel mutagenesis strategy combining point and structural mutagenesis and constructed yeast with high 2,3-BDO tolerance. The strategy developed in this study achieves efficient modification of yeast genomic DNA, and thus it can be applied for construction of mutant yeasts with various stress tolerance. Besides, it is expected to further elucidate resistance mechanisms and further improve tolerance by detailed analysis of mutants with improved tolerance.
Acknowledgments
This work was partly supported by JSPS KAKENHI (grant number JP18K14069 and JP18KK0413).
Reference
1. H. Abe et al.; J Biosci Bioeng 108:199–204 (2009)
Isopentenol is promising biofuel with more favorable combustion properties. Because isopentenol shows some attractive characteristics, such as higher energy density, lower water miscibility, and better low-temperature fluidity than other biofuels, it is expected as alternatives of gasoline and jet-fuel. Supplies of petroleum, the conventional source of isopentenol, are unsustainable, and chemical synthesis processes could cause serious environmental problems. As an alternative, the biosynthesis of isopentenol from biomass is more sustainable and environmentally friendly. Therefore, we tried to produce isopentenol by engeneered Escherichia coli. Firstly, we introduced seven genes (atoB, mvaS, mvaE, ERG12, ERG8, MVD1 and nudB) into Escherichia coli and used this engineered strain to produce isopentenol from glucose. This strain (ISO-1) produced 68.1 mg/L isopentenol and accumulated 3.83 g/L mevalonate from 20 g/L glucose. Secondly, we constructed ISO-2 strain enhanced pathway after mevalonate. This strain produced 171 mg/L isopentenol and accumulated 1.01 g/L mevalonate from 20 g/L glucose. Thirdly, we constructed ISO-2 with glcB knockout (ISO-2ホ・I>glcB) and aceBA knockout (ISO-2ホ・I>aceBA) and tried to strengthen the flux to isopentenol. As a result, it succeeded in further improving the isopentenol production. ISO-2ホ・I>glcB and ISO-2ホ・I>aceBA produced 434 mg / L and 472 mg/L isopentenol. This result suggests that reducing Acetyl-CoA consumption and redirecting metabolic flux for isopentenol production is important for isopentenol production. ISO-2ホ・I>aceBA succeeded in about 7 times isopentenol production of ISO-1.
Fatty acids are used for fuels, cosmetics, pharmaceuticals. Currently, they are made from natural fats and oils. Production technology using bacteria has been studied, but it has not reached a practical level. Previous studies have shown that palmitic acid and oleic acid account for 90% of the fatty acids excreted from cells when producing fatty acids from glucose using Corynebacterium glutamicum. In this study, we tried to produce palmitic acid and oleic acid using metabolically engineered C. glutamicum. First, we disrupted fasR(strain1). Second, we introduced the plasmid containing cgR0949-tfu0939 encoding cellobiose degrading enzyme, β-glucosidase(BGL), into strain1 to produce fatty acid from cellobiose. This strain intracellularly produced 0.606 g/L of palmitic acid and 0.536 g/L of oleic acid from cellobiose in test-tube after 72h. Third, we disrupted two genes (pyc and ppc) to accumulate acetyl-CoA. The strain disrupted fasR and pyc (strain2) extracellularly produced 0.130 g/L of palmitic acid and the strain disrupted fasR and ppc (strain3) ectracellularly produced 0.0676 g/L of oleic acid from glucose in the test-tube after 72h.
Pichia membranifaciens KS47-1 have been isolated from environment as a resistant yeast as growth inhibitory materials in hydrolysates derived from lignocellulosic biomass. The strain has a great potential of host yeast for cell factories to be applied a broad ranges of bio-products, after introducing metabolic pathways. In this study, the responses of gene expression in the strain were differentially analyzed between in a corncob hydrolysate and in the hydrolysate removing the inhibitory materials by activated carbon treatment by RNA-sequence analysis. The hydrorated corncob (1:10 of solid/water ratio) were hydrolyzed at 121°C for 1 h by 3.0 % sulfuric acid, and subsequently treated at 50°C for 3 days by a cellulase. Activated carbon (AC) of 3% w/v were added and absorbed inhibitory materials in the hydrolyastes for 3 h, and were removed by a filteration. Although KS47-1 growth was observed in the both of hydrolysates prepared, the latent growth was shown in the hydrolyastes without AC treatment. Duplicated RNA-seq analyses were performed for the logarithmic growing cells in each hydrolysates with 11.4 to 13.7 M reads (Q30: >94%). Top-Hat2 mapping was carried out with 103% of mapping ratio and 30% coverage. The sequencing depths were among 4.4 to 6.8 folds. According to gene ontology analysis, toxicity compounds in hydrolysates stimulated many of membrane transporters. The expression levels of ATP-binding multi drag transporters, TPO3, SNQ2, and PDR5 were increased 1.74, 1.63, and 1.46 as log2 fold changes, respectively (p value = 0.00005). The results implied that KS47-1 efficiently evacuate the inhibitory compounds by the activities of the transporters responded to increasing the toxic compounds in the cytosol.
In order to solve environmental and resource exhaustion problems and achieve an environmental sustainable society, a biorefinery that produces a variety of bio-based products from renewable biomass resources is important. At present, by microbial organisms using C5/C6 sugars, the production of target compounds currently being produced from petroleum is important. In particular, it is expected that the demand for unsaturated hydrocarbons having double bonds, which can be used as a polymer raw material, will increase rapidly.
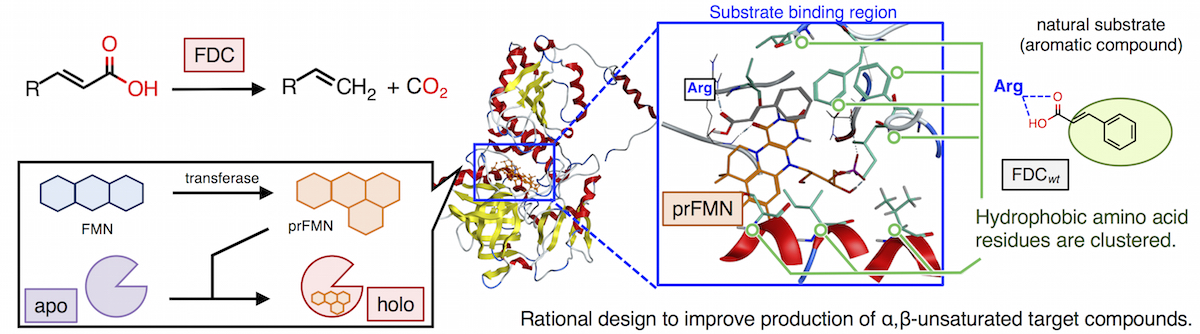
To produce these compounds, we focused on ferulic acid decarboxylase (FDC). FDC can catalyze the decarboxylation of the R-C=C-COOH side chain end of the hydrocarbon unsaturated carboxylic acid (such as ferulic acid) and converts it to R-C=C (such as styrene). For this decarboxylation reaction, FDC requiters a prenylated-FMN (prFMN) as a cofactor, which is synthesized from FMN and dimethylallyl-phosphate by prenyltransferase.
In this study, to bio-synthesize target α,β-unsaturated compounds, we screened several genes encoding FDC and prenyltransferase. Then the FDC showed the highest decarboxylase activity was selected as a template and we developed FDC variants to enable production of target molecules efficiently based on the rational design of enzyme. Recent years has seen significant interest in a rational design in which a mutation is introduced at pinpoint relative to the active site of the enzyme. As a host for enzyme screening and substance production, Escherichia Coli was selected. Finally, with the combination of precursor-producing microbial cell and decarboxylase mutants, we aim to develop a microbial cell factory that produces α,β-unsaturated target compounds efficiently and directly from renewable carbon source glucose.
Metabolic engineering is a promising method for maximizing the production of specific metabolites. Saccharomyces cerevisiae and Escherichia coli are common hosts for metabolic engineering. However, these microorganisms require an aerobic growth phase with aeration and vigorous agitation before the fermentation production phase. As a consequence, bioproduction systems are often economically unsustainable, particularly for the bulk production of chemicals or biofuels. Moreover, because of the requirement for aeration during the growth phase, it is usually challenging to scale-up the fermentation process. This study aimed to construct a platform microorganism for anaerobic bioprocessing (ABP), which requires neither aeration nor vigorous agitation.
The lactic acid bacterium Lactobacillus plantarum maintains a similar growth rate under both aerobic and anaerobic conditions, which enables the growth-associated production of lactate without the need for aeration and vigorous agitation. Therefore, L. plantarum is a suitable host for ABP. However, the strain converts most of the fermented sugar into lactate. To prevent the production of lactate, we disrupted both L- and D-lactate dehydrogenase genes (ldhL1 and ldhD, respectively) in L. plantarum. Then, to promote ethanol fermentation, we introduced in the ホ・I>ldhL1 ホ・I>ldhD strain pyruvate decarboxylase and alcohol dehydrogenase genes from various origins. For the strain showing the highest ethanol titer, ethanol fermentation was performed from 150 g/L glucose in a 1 L jar fermenter. After 100 h of fermentation, 56.7 g/L of ethanol was successfully produced.
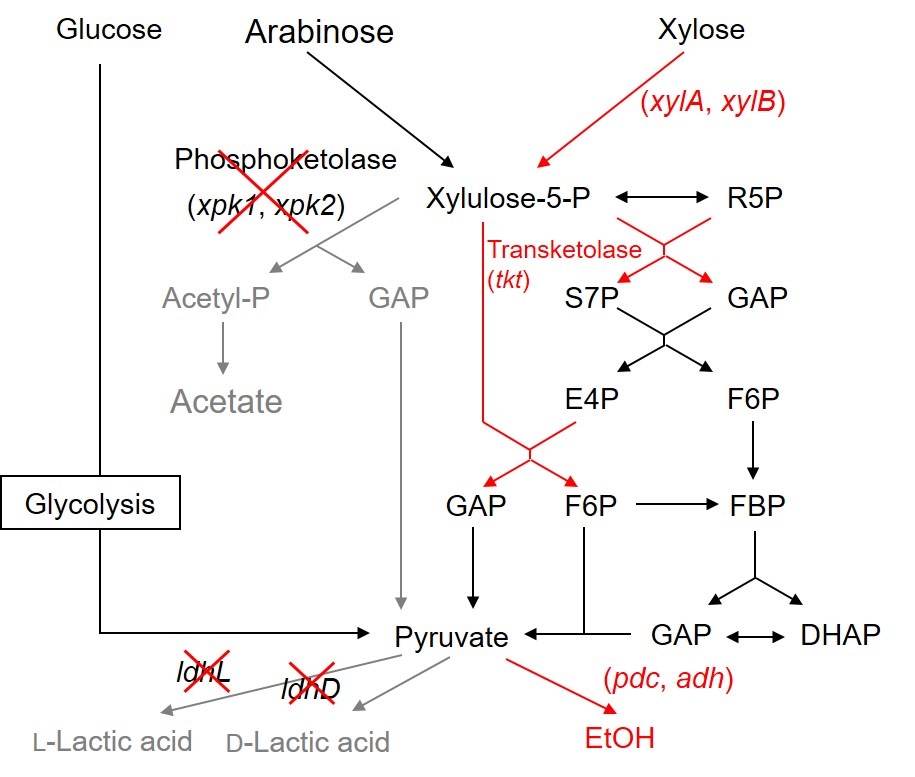
Our next challenge is to produce ethanol from lignocellulose hydrolysates which contain both C6 and C5 sugars. To redirect the phosphoketolase pathway (which produces equimolar amounts of pyruvate and acetate from pentose sugars) to the pentose phosphate pathway (which only produces pyruvate from pentose sugars), we replaced the phosphoketolase gene (xpk1) with the transketolase gene (tkt) from Lactococcus lactis. Using this strain, ethanol fermentation from C5 sugars are now under consideration.
1. Introduction
Polyhydroxyalkanoates (PHAs) are biodegradable and produced by microorganisms from biomass. They are expected to reduce plastic wastes and carbon dioxide emissions compared to conventional fossil resource-derived plastics. Compostable PHA filtration membranes will be useful to develop environmentally benign filtration processes. In this study, PHA porous membranes were prepared using phase separation method, and membrane forming conditions were examined for application to microfiltration membranes.
2. Materials and methods
Typically PHA (extrusion grade, Goodfellow) was dissolved at 10-20% on a hot stirrer at 85 °C in 1,4-dioxane (DIOX), 1-methyl-2-pyrrolidone (NMP), or N,N,-dimethylformamide (DMF). After cooling to 50 °C, the PHA solution was cast onto a glass plate with a 0.5 mm thick frame which was preheated at the same temperature as the solution. It was immersed in a water bath (25 °C) to induce its phase separation. The filtration resistances of formed flat-sheet membranes were calculated from the filtration rates measured by the filtrations of purified water at 10 kPa, and the retention ratios were from the filtrate turbidity at a wavelength of 660 nm after filtration of suspensions of latex beads with a diameter of 0.3 ホシm.
3. Results and discussion
Flat sheet membranes were obtained from the 20% PHA solutions while no flat ones were obtained from the 10% polymer solutions. The prepared PHA flat membranes have asymmetric structures having a dense layer in the surface portion in contact with non-solvent water at the time of phase separation, and a porous structure inside. The PHA membrane prepared with DMF as a solvent showed the lower filtration resistance and the higher retention of latex beads compared to the membrane prepared with DIOX and NMP. The polymer concentration was optimized to be 18%. The PHA filtration membrane will be one of the key materials to achieve sustainable development goals (SDGs).
Fatty acids are valuable products because they have wide industrial applications in the manufacture of detergents, cosmetics, food, and various biomedical applications. In enzymecatalyzed hydrolysis, the use of immobilized lipase results in high production cost. To address this problem, Eversa Transform lipase, a new and low-cost liquid lipase formulation, was used for the first time in oil hydrolysis with gac oil as a triglyceride source in this study. Response surface methodology was employed to optimize the reaction conditions and establish a reliable mathematical model for predicting hydrolysis yield. A maximal yield of 94.16% was obtained at a water-to-oil molar ratio of 12.79:1, reaction temperature of 38.9 degree C, enzyme loading of 13.88%, and reaction time of 8.41 h. Under this optimal reaction condition, Eversa Transform lipase could be reused for up to eight cycles without significant loss in enzyme activity. This study indicates that the use of liquid Eversa Transform lipase in enzyme-catalyzed oil hydrolysis could be a promising and cheap method of fatty acid production.
Rice husk, a form of biomass material, is a potential source of renewable energy. Recovering silica from rice husk is an important element in manufacturing high value-added products, and ensures efficient utilization of a bio-resource. This paper reports the synthesis of SBA-15 mesoporous silica from recycled rice husk ash waste. Next, graphene oxide/order mesoporous carbon (GO/CMK-3) nanocomposite was synthesized using the SBA-15 template. This investigation characterized GO/CMK-3 materials through transmission electron microscopy (TEM), field–emission scanning electron microscopy (FESEM), X-ray diffractometry (XRD), surface area analysis, and Raman spectrometry. Various experimental parameters including the kind of adsorbent, initial concentration of dye, and solution temperature were investigated. TEM of the synthesized GO/CMK-3 revealed that carbon material possessed a well-ordered array of mesopores with a uniform pore size. The surface area and pore volume of RH-CMK-3/GO sample were 936 m2/g and 1.077 cm3/g, respectively. GO/CMK-3 showed the higher capacity for dye adsorption than that of pure SBA-15 and CMK-3. Increasing the initial concentration of methylene blue (MB) could enhance the adsorption rate. The adsorption capacities were decreased with an increase in the solution temperature. The thermodynamic analysis reveals that the adsorption process was exothermic and spontaneous. The GO/CMK-3 composite contained considerable degree of oxidation with different functional groups, which could efficiently bind a dye molecule to form a dye complex. The GO/CMK-3 composite is a potential low-cost adsorbent for removal and recovery of industrial dye from wastewater.
Shochu lees is a distillation residue discharged during shochu production, and it is one of major organic wastes available for sustainable bioenergy production. The nonconformity with the composition variation of the waste may lower the activity of methanogens in a two-stage hydrogen and methane fermentation process. One of them is the effect of organic acid concentration on methane production. In order to maintain the activity of methanogens, the organic acid concentration in the substrate must be kept constant. However, the appropriate organic acid concentration for the activity of methanogens has not been reported. In methane production, 70% of the produced methane is due to the decomposition of acetic acid and is carried out by the acetylotrophic methanogens. In this study, we focused on acetic acid concentration. Sodium acetate (0.002, 0.01, 0.02, 0.03 and 0.2 mol/L) was added to the fermented broth which had been acclimated for a long time, and cultivation was carried out at 37 °C, at pH 7. Methane gas was most generated at acetic acid concentration of 0.03 mol/L. Therefore, it was found that the acetic acid concentration in the substrate of 0.03 mol/L was suitable for the activity of acetylotrophic methanogens in methane production. The results showed that the activity of methanogens can be maintained by maintaining the acetic acid concentration at 0.03 mol/L in the acetic acid concentration in the substrate. In this case, methane was produced immediately after the start of fermentation. This study showed that by maintaining the acetic acid concentration in the substrate at 0.03 mol/L, the activity of methanogens is maintained and the production of methane in a short time can be achieved.
Introduction
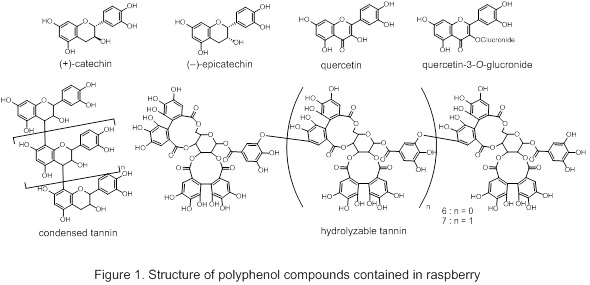
It is known that red raspberry (Rubus idaeus) contains various polyphenol compounds at high concentration and its functionality is expected to have health maintenance and disease prevention effect. Not only the fruits, but also the leaves of raspberry plants, are highly esteemed for tea making around the world and are largely used for food. We are interested in the relationship between the polyphenol content of raspberry and the cultivation environment. In this report, we discuss the results of on the effect of light and temperature on polyphenol accumulation in raspberry leaves.
Results and discussion
When raspberry was cultivated in a plant factory unit and light intensity, wavelength, and temperature were varied, the amount of total polyphenol increased under blue light. Quantitative determination of flavan-3-ol derivatives {(+)-catechin, (–)-epicatechin and condensed tannin}, quercetin derivatives and hydrolysable tannin (Figure 1), was carried out using HPLC. Among them we confirmed flavan-3-ol derivatives increase under blue light. Semi-quantitative RT-PCR showed no correlation between flavan-3-ol derivatives-related gene expression and the amounts of the compounds measured in the leaves.1
Ref. (1) R. Kobori et al., Metablites,2019,9, 56.
Cattle urine is one of large amount of waste in dairy industries, and often insufficiently decomposed in a reservoir and sprayed on farmland as fertilizers. However, the immature fertilizers sprayed causes problems in exhausting awful smell and in water pollutions of land water. In this study, it investigated the botanical effects of mature and immature cattle urine on germination of Japanese mustard spinach, Brassica rapa var. perviridis, and enhancing growth of tomato, Solanum lycoperisicum L., and B. rapa. Treatment by long-term aerations led microbial degradation of malodorous substances and germinating-inhibitory materials, and reduced the awful smell from the fermented urine. Immature fermented urine inhibited germination of B. rapa, but mature urine never. In hydroponics, the growth of B. rapa and S. lycoperisicum were enhanced by the matured fermented urine, with diluting 1/10 in water (Fig. 1). Equivalent results were observed in a commercial culture soil. Murashige and Skoog medium including 1/10 mature fermented urine also enhanced the growth of B. rapa. The weight with the mature fermented urine was three fold larger than that without the fermented urine. Total nitrogen, phosphate and potassium as inorganic nutrients were less than 290 mg-N/L, 26 mg/L, and 2,700 mg/L, respectively. The amounts of N, P, and K as nutrients were insufficient to stimulate the growth to the equivalent levels, as inorganic fertilizers. Interestingly, the fermented urine maintained the growth stimulating ability after autoclaving (121 °C, 20 min). From the results, there is possibility that thermal-stable organic chemicals in the sufficient fermented urine can act to stimulate the growth of plants, and next-generation fertilizers from cow urine may be produced in the future.

The utilization of biomass is required to establish a sustainable society. Rice bran, a byproduct in the refining of brown rice, is mainly used for producing rice oil. At this time, the defatted rice bran is generated as a residue. A method of effectively utilizing defatted rice bran is required. Several studies have reported that defatted rice bran contains various functional substances, especially protein and amino acids. The protein of rice bran has a highly nutritious with a well-balanced amino acid composition. Rice bran also contains many useful amino acids, such as γ-aminobutyric acid which has various functionalities.
We suggested that the preparation of an extract containing functional substances from defatted rice bran could be connected to new possible applications. Hot compressed water was used as the solvent for extraction. We expected that hot compressed water would hydrolyze the protein of defatted rice bran and increase the amount of amino acid extracted. The purposes of this study are to evaluate amino acids in the extract prepared from defatted rice bran and to reveal the reaction of protein or amino acids in hot compressed water.
The defatted rice bran was prepared from rice bran by the extraction process using hexane. The defatted rice bran was treated under different temperature conditions to prepare an extract. The extract was deproteinized and subjected to amino acid analysis.
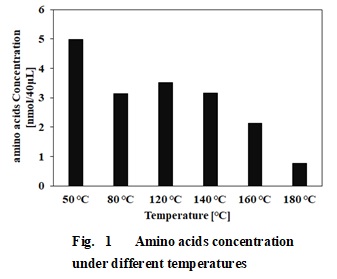
Fig. 1 shows the total amount of 20 amino acids that constitute a protein obtained at each of the evaluated temperatures. The total amount of amino acids decreased as the temperature increased. The results suggest that the protein of rice bran was not decomposed amino acids in these temperatures. It is also considered that amino acids were decomposed or bonded to other substances by intermolecular dehydration or Maillard reaction.
The production of biofuels by engineered microorganisms has recently become an attractive option, because of the global environmental problems and the combustion of fossil fuels. Among various biofuels, long chain alkanes and alkenes are superior compared with other biofuels like ethanol due to its high energy density and compatibility with modern internal combustion engines. Although an Escherichia coli strain that produces alkanes and alkenes was constructed by heterologous expression of acyl-acyl carrier protein reductase from Synechococcus elongates (SeAAR) and aldehyde decarbonylase from Nostoc punctiforme (NpAD) (1), its productivity did not fulfil industrial requirements.
In this study, effects of the promoter for NpAD expression on the alkanes production by engineered E. coli was evaluated. And improvement of the solubility of SeAAR was tried.
Plasmids pESeAAR-trcNpAD and pET7SeAAR-T7NpAD which express SeAAR and NpAD simultaneously were constructed. pESeAAR-trcNpAD expresses SeAAR by T7 promoter and NpAD by trc promoter, whereas pET7SeAAR-T7NpAD expresses both SeAAR and NpAD by T7 promoter. E. coli BL21 (DE3) cells were transformed using these plasmids and the resultant transformants were named BL21/pESeAAR-trcNpAD and BL21/ pET7SeAAR-T7NpAD. Furthermore, random mutation was introduced into the SeAAR gene using error-prone PCR to obtain mutated SeAAR which solubility was improved.
The amounts of alkanes produced by BL21/pET7SeAAR-T7NpAD was 1.5-fold larger than those by BL21/pESeAAR-trcNpAD. And mutated SeAAR which solubility was improved was successfully obtained.
(1) Andreas, S. et al.; Science, 329, 559-562 (2010).
We successfully isolated new freeze-tolerant yeast SRT88 from Hokkaido Saroma Lake. In this study, taxonomic characterization of SRT88 and bread manufacturing using the strain. The strain identified as Zygosaccharomyces rouxii that is known as a fermenting yeast in soy source manufacturing. The freeze tolerance of the yeast was observed higher than that of Torulaspora delbrueckii NBRC1129 known as freeze tolerant yeast. To characterize the strain in the dough for frozen storage method, fermentabilities in dough were examined after freezing and thawing. Although SRT88 fermentability demonstrated 117 mL lower than a commercial dry yeast 316 mL, the strain showed 114 mL after 1 week-frozen-storage which was higher fermentability than a dry yeast of 67 mL and maintained the level of fermentability after 2 weeks-frozen-storage. Furthermore, SRT88 fermentability was improved to use glucose. The fermentability using glucose was 40% higher than that using sucrose. In an analogous fashion, crumb hardness of baked bread using SRT88 was 1.17ツア 0.16 N , on the other hand after storage was 1.68ツア0.23 N lower than that using a commercial dry yeast was 1.92ツア0.40 N. According to the results, SRT88 showed an advantage for bread manufacturing including frozen storage. SRT88 gave different flavor to bread from commercial dry yeast on the basis of volatile component analysis with GC/MS and statistical analysis of orthogonal projections to latent structures discriminant analysis (OPLS-DA). Heat map result showed that SRT88's bread has characteristic volatile component of 2-Ethyl-1-hexanol (Fig. 1).
In conclusion, SRT88 showed stable fermentability during frozen dough method and flavor is different from commercial dry yeast to fewer frozen damage to dough after frozen. The results demonstrated a possibility that SRT88 use frozen dough method

Non-conventional yeast (NCY) in cool temperate zone and subarctic zone has possibilities of adapting to frozen stress in winter. Several freezing-tolerant-NCYs, which demonstrated higher freezing-resistance than previous reported yeasts, have been isolated from environment. The mechanisms of frozen resistance of the isolates have not been revealed. Saccharomyces cerevisiae, well-known yeast species, accumulates sugar alcohols and amino acids in their cytosol, and resists to frozen stresses with buffering by the materials. In this study, we investigated the frozen-adapting mechanism of the isolates, Zygosaccharomyces rouxii SRT88 and Candida tropicalis SRF61, to comparing metabolomes among the taxonomically related strains.
Metabolites were extracted by methanol/chloroform (1:2) from freeze-dried cells, and trymethylsilirated by methoxamine hydroxylate and N-Methyl-N-trimethylsilyltrifluoroacetamide. The trimethylsilylated derivatives were analyzed by GC-MS. Orthogonal projections to latent structures (OPLS) were used for modeling the differences of metabolites.
According to OPLS, high correlated modeling were achieved between metabolite composition and freeze-resistance (R2>0.95, Q2>0.90) .The significant metabolites were filtered by variable importance in projection (VIP) scores and p values in analysis of variance (VIP ≥ 1.0, p ≤ 0.05).Based on the results, C. tropicalis SRF61 accumulated higher concentrations trehalose and aspartic acid than the referenced strains, and Z. rouxii SRT88 higher D-mannitol. Aspartic acid was a novel cryoprotectant metabolite in the best of our knowledge. The possibility of variety in freezing-tolerant mechanism of NCYs were demonstrated in this study.
Microbial production of 1,5-diaminopentane is from renewable feedstock is a promising and sustainable approach to produce bio-based polyamides. In this study, we constructed beta-glucosidase (BGL) secreting Corynebacterium glutamicum, and direct 1,5-diaminopentane production from cellobiose as well as glucose was demonstrated. First, C. glutamicum was metabolically engineered to create L-lysine (a direct precursor of 1,5-diaminopentane) producing strains. Then L-lysine decarboxylase (ldcC) derived from E. coli and BGL from Thermobifida fusca YX (Tfu0937) were co-expressed. This engineered C. glutamicum strain produced 27 g/L of 1,5-diaminopentane from cellobiose in CGXII minimal medium in a fed窶臣atch cultivation. The yield was 0.43 g-1,5-diaminopentane / g-glucose (1 g of cellobiose corresponds to 1.1 g of glucose). These results demonstrated the feasibility of 1,5-diaminopentane production from cellobiose or cellooligosaccharides as alternative substrates using engineered C. glutamicum strain.
Mevalonate is a precursor of isoprenoids and has been used in cosmetics and as building block for the production of functional polymers. It is difficult to industrially produce mevalonate by chemical synthesis, because mevalonate used in cosmetics is need to be (R)-enantiomer. Thus, mevalonate is produced by fermentation of microbes from glucose. However, there still remain problems that yields of mevalonate is not sufficient. Therefore, we tried to develop highly efficient production of mevalonate by metabolic engineering of Escherichia coli.
First, we introduced into E.coli three genes which express Acetyl-CoA acetyl transferase, HMG-CoA synthase and HMG-CoA reductase (atoB, mvaS and mvaE respectively). This strain produced 2.16 g/L mevalonate from 20 g/L glucose.
Next, we tried to improve production capacity of mevalonate by disrupting some genes. Because three molecules of acetyl-CoA are required to synthesize one molecule of mevalonate, the amount of acetyl-CoA is considered to affect the production of mevalonate. To accumulate acetyl-CoA, we constructed MG1655ホ廃pcホ蚤ceBA strain. This strain produced 6.00 g/L mevalonate from 20 g/L glucose.
Finally, we tried to produce mevalonate from cellobiose. To give the E.coli an ability to break down cellobiose, we displayed β-Glucosidase(BGL), Tfu0937, on the E.coli cell surface using Blc as an anchor protein. BGL-displaying MG1655ホ廃pcホ蚤ceBA strain produced 5.72 g/L mevalonate from 20 g/L cellobiose. We succeeded in producing mevalonate from cellobiose for the first time.
Shikimic acid is a valuable hydroaromatic compound and a key metabolic intermediate. Currently, shikimic acid can be obtained from fruit of Chinese star anise, but there are problems of feedstock limitation and extraction cost. Therefore, production of shikimic acid from renewable resources such as cellulosic biomass is required. Corynebacterium glutamicum is widely used for industrial production of amino acids including L-glutamate and L-lysine. However, C. glutamicum doesn't accumulate shikimic acid, and can not assimilate cellulose originally. Cellulose is degraded to cellobiose by cellulolytic enzymes, and cellobiose is degraded to glucose by β-glucosidase(BGL). At first, we tried to produce shikimic acid from cellobiose using metabolically engineered C. glutamicum with BGL. In this study, to accumulate shikimic acid in C. glutamicum, we disrupted several genes including aroK, qsuD, qsuB, nagD. We also disrupted pyk gene for improving availability of PEP, and mutated gnd gene to improve pentose phosphate pathway. Furthermore, we expressed the BGL gene(cgR0949-tfu0939) in C. glutamicum to produce shikimic acid from cellobiose. As a result, we successfully produced 2.8 g/L of shikimic aicd from cellobiose using engineered C. glutamicum. Deletion of pyk and mutation of gnd improved shikimic acid productivity.
Microbial bioproduction have attracted much attention because of the increasing demand for environmentally-friendly bioprocesses. We focused on the production of mevalonate by Escherichia coli. Mevalonate is one of the important compounds, which is utilized as the intermediate of a medicinal or a cosmetic compound and is the metabolic intermediate for synthesizing terpenoids. Here, we attempt to improve the production of mevalonate by using single gene knockout strains. The plasmid including mevalonate-producing genes (pMev) was transformed into a single gene-deficient E. coli BW25113 strain, and cultivated with 20 g L-1 glucose and 5 g L-1 yeast extract containing minimal medium. Mevalonate production was determined using a gas chromatograph equipped with a flame ionization detector. BW25113 harboring pMev produced mevalonate from glucose at a concentration of 0.8 g L-1 after 48 hours cultivations (WT). Then, the effect of single gene knockout on mevalonate production was evaluated. The deletion of several genes was effective for improving the production of mevalonate. The production of mevalonate with a gltA-deficient strain, an aceA-deficient strain, an aceB-deficient strain and an acnA-deficient strain were increased compared with WT (1.4 g L-1, 0.9 g L-1, 1.3 g L-1, 1.0 g L-1, respectively). These genes are involved in acetyl-CoA catabolism in the glyoxylate shunt, and acetyl-CoA level is critical for enhancing the production of mevalonate because acetyl-CoA is metabolic intermediate for the synthesis of mevalonate. The deletion of cyaA, which is involved in catabolite repression, was also effective for the improvement of the production of mevalonate (1.7 g L-1).
Photosynthetic microalgae attract much attention as a sustainable green cell factory, and genetically modified microalgae have been expected to be a useful tool for bioenergy and recombinant protein production. However, random integration of transgene in the microalgae nuclear genome is susceptible to gene silencing of heterologous gene expression1. Here, we attempted to perform targeted gene integration into a pre-determined nuclear genomic site of Chlamydomonas reinhardtii using Cre/loxP recombination system for stable transgene expression. We constructed an expression vector plasmid encoding reporter genes (zeocin resistant gene and green fluorescent protein [GFP] gene; Zeo-2A-GFP) and mutated loxP to generate founder cells. The plasmids were introduced into C. reinhardtii by electroporation and the cells were screened by zeocin. Cell clones exhibiting the stable expression of the reporter genes during culture were selected from transformants using FACS. A donor vector containing target genes flanked by corresponding mutated loxPs was constructed and introduced into founder cells together with a Cre expression vector. After Cre-mediated gene integration, paromomycin resistant gene and IFNα-4 expression cassettes of the donor vector were integrated into a pre-introduced target loxP site placed downstream a promoter. Targeted knock-in cells lost the zeocin resistance but acquired paromomycin resistance. The optimal ratio of donor vector and Cre expression vector was determined by counting the number of paromomycin resistant colonies. For the established clones, the targeted integration was confirmed by genomic PCR using various specific primer sets. RT-PCR revealed that IFNα-4 was expressed in five independent transgenic cell lines tested. In conclusion, we succeeded in site-specific transgene integration into the C. reinhardtii cell genome using Cre/loxP system. This result suggests that Cre-based cell engineering is a promising approach to generate smart microalgae expressing foreign genes.
1. Scranton MA et al., Plant J. 82(3):523-531 (2015)
Chitosan and its derivatives have been widely used as an antimicrobial agent either alone or blended with other polymers. The chitosan derivatives with quaternary ammonium groups have higher efficient activity against bacteria as compared to pure chitosan, such as N-[2-hydroxyl-3-trimetylammonium) propyl] chitosan chloride (HTCC). However, HTCC is a water soluble derivative of chitosan. Hence, biodegradable polyvinyl alcohol (PVA) was chosen and used as an effective additive to HTCC to improve fiber spinning ability. In this work HTCC/polyvinyl alcohol nanofiber membrane has been successfully made by electrospinning process. The HTCC/PVA nanofiber can be stabilized against water by isothermal crosslinking with water soluble blocked di-isocyanate. Microbiological assessments showed that the water stable HTCC nanofibers have a good antibacterial activity against E coli.
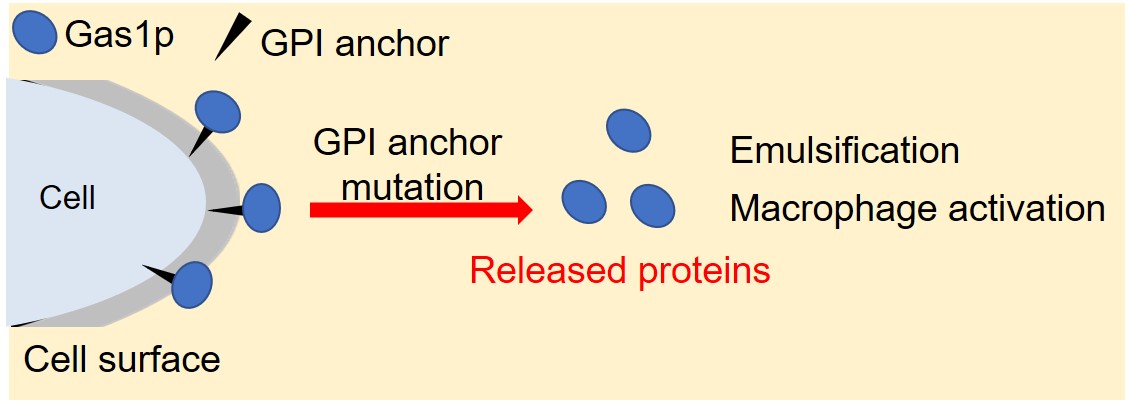
Emulsifiers that uniformly mix immiscible materials such as water and oil are widely used in food, cosmetics, and so on. Although many emulsifiers currently used are chemically synthesized, naturally-occurring emulsifiers are required from the viewpoint of the safety sought by consumers. We found that the cell wall mutant of Saccharomyces cerevisiae widely used in fermented food had a strong emulsifying effect on the cells themselves. Therefore, we focused on the emulsifying effect of cell wall proteins. Then, we found high emulsifying activity in Gas1 protein, which is bound to the cell wall via glycosylphosphatidylinositol (GPI) anchor. In addition, it was found that the Gas1 protein strongly activates mouse macrophage, the immunocompetent cell. Thus, this protein is expected to be applied as an emulsifier capable of imparting an immune enhancing effect. Gas1 protein belongs to Gas family of proteins in S. cerevisiae, including other four proteins with high homology. When we examined the relationship between protein structure and their activities by evaluating other Gas family proteins and Gas1 mutant proteins, the region required for high emulsifying activity was elucidated. Currently we are clarifying the regions required for macrophage activation. Meanwhile, S. cerevisiae has many proteins localized in the cell wall via different bond modes from GPI anchor. We have been also examining emulsification and macrophage activation for such proteins, and our results suggest that several proteins have the same function as Gas1. We think that there are proteins involved in emulsification and macrophage activation in the cell surface of S. cerevisiae and we will surely develop not only naturally-derived emulsifiers but also functional emulsifiers by utilizing them. This presentation will report the above details.
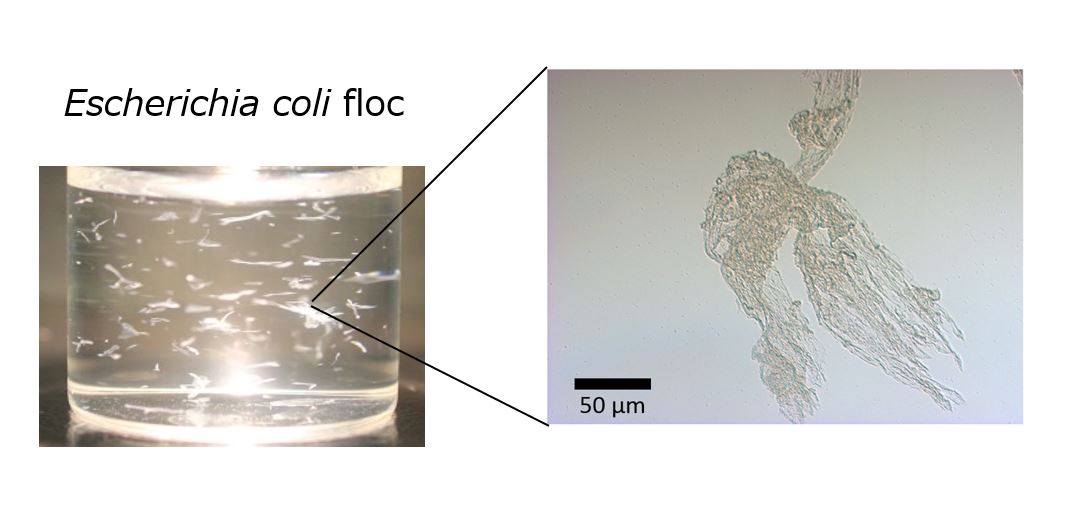
Flocculation is an aggregation phenomenon of microbial cells in which the cells form flocs. Flocculation can contribute to the efficiency of industrial production processes. For example, in repeated batch fermentation production of ethanol using flocculating yeast, the centrifugation step required for cell recovery can be eliminated. Besides yeast, Escherichia coli is known as an industrial bacterium. Although E. coli naturally does not form floc, we succeeded in inducing them to floc formation by overexpression a native bcsB gene, encodes a component of cellulose synthase or deletion of degP gene, encodes a periplasmic protease1). However, floc formation is less than 100 mg/L and not enough to be used in fermentation production. In this study, we aimed to understand the formation mechanism of E. coli flocs and further increase the formation efficiency. Since E. coli flocs were proteinous, the amount of protein included in flocs were measured, and the value was used as an index of floc amount. When each strain was cultured in LB medium containing glycerol, only the ホ・I>degP strain increased the floc formation. Amount of floc formation reached approximately 230 mg/L in the presence glycerol. Meanwhile, the uptake of glycerol into cells was not confirmed. Next, we focused on the high viscosity of glycerol. Additing thickening agent such as carboxymethyl cellulose and polyvinylpyrrolidone into medium did not increase the floc formation. In contrast, the addition of ethylene glycol, which is classified into the same polyols as glycerol, significantly promoted the floc formation. Thus, it was suggested that floc formation of E. coli ホ・I>degP strain was enhanced by polyols. Further analysis will aim to elucidate the action of polyols on E. coli flocs.
1) Ojima et al. (2015) AEM 81:5900-5906
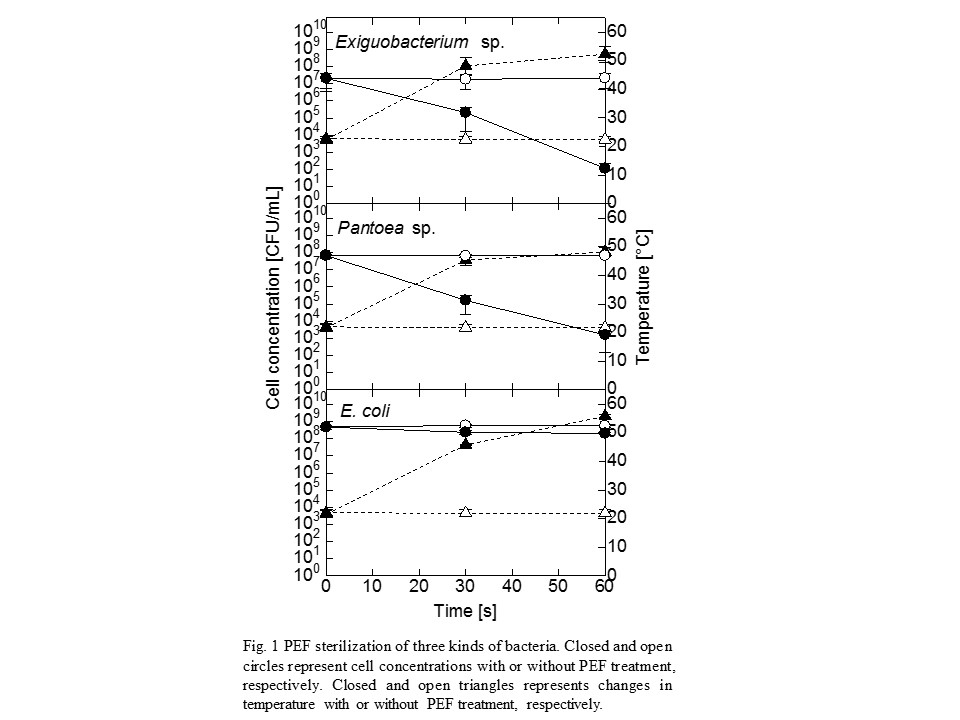
From the standpoint of the food safety management, it is very important to control microorganisms that can grow in a refrigerator. We evaluated the growth characteristics and sterilization conditions of Exiguobacterium sp. and Pantoea sp. which were isolated from a food manufacturing environment. These two microorganisms were capable of growth at refrigeration temperatures (4 °C and 10 °C), but their optimal growth temperature was approximately 30 °C. Thus, these microorganisms were classified as psychrotrophic bacteria. Heat treatment and sodium hypochlorite exposure were effective for the inactivation of the two microorganisms and the sensitivities toward these sterilization conditions of them were higher than those of Escherichia coli (E. coli). Although Exiguobacterium sp. and Pantoea sp. exhibited similar alkaline tolerance at pH 8.0, the degrees of their acid tolerance were different from each other. Exiguobacterium sp. was easily killed by acetic acid (pH 2.68) and citric acid (pH 2.0), while Pantoea sp. was relatively insensitive to these acids. In particular, Exiguobacterium sp. should be careful because the microorganism exhibited an unusually high resistance to ozone treatment and UV radiation whose conditions were enough to kill Pantoea sp. and E. coli. This is probably due to the protective effects of carotenoids, antioxidant enzymes and DNA repair enzymes in Exiguobacterium sp. cells against oxidative stress and DNA damage. Interestingly, Exiguobacterium sp. and Pantoea sp. were effectively sterilized by pulsed electric field (PEF) treatment under the condition that was not sufficient to kill E. coli (Fig. 1). Since the Exiguobacterium sp. and Pantoea sp. cells were cultivated at 4 °C, the cell membranes of the two microorganisms seemed to become more fragile with increasing unsaturated fatty acid content. Our results suggested that PEF treatment has a high potential to be used in the sterilization of psychrotrophic bacteria associated with spoilage of food at refrigeration temperatures.
Beta-carotene is distinguished as a vitamin A precursor and make an effect of the antioxidant action and immunostimulating activity. In recent years, it is mainly produced by extraction from plants and chemical synthesis from petroleum which causes an unstable production and serious environmental problems. In order to achieve the requirements of mass production, in this study, we carried out metabolic engineering using CRISPR-Cas9 to edit genes of beta窶芯arotene biosynthesis from cellulosic biomass in Schizosaccharomyces pombe. This yeast is superior in protein production capacity compared to other yeasts. First, we carried out destruct genes of encoding proteases and acetaldehyde dehydrogenase. At the same time, we introduced carotenogenic genes (crtYB, crtE, crtI) into genomic DNA of S. pombe. And this recombinant strain is successfully produced the beta-carotene from glucose. Furthermore, a main component of cellulosic biomass is cellulose. Cellulose is degraded to cellobiose by cellulolytic enzymes and cellobiose is degraded to glucose by beta-glucosidase (BGL). To produce beta-carotene from cellobiose, BGL gene is introduced into S. pombe and expressed BGL on cell surface. As a result, this recombinant strain is successfully produced 1864 μg / g DCW beta-carotene from cellobiose. It is assumed that adjusting the Redox balance and strengthening metabolic flux enhance beta-carotene production.
Cordycepin, a nucleoside analogue (3'-deoxyadenosine), is the major active constituent of Cordyceps militaris and was first reported as a metabolite isolated from a culture broth of C. militaris. To date, many biological functions of cordycepin have been discovered; cordycepin has anti-tumor, anti-metastatic, anti-bacterial, anti-viral, immunomodulatory and anti-inflammatory activities. In the last decade, studies targeting cordycepin as a therapeutic agent have been in progress. These studies include application to a variety of cancers, especially leukemia (ClinicalTrials.gov, verified by OncoVista, Inc., 2009), trypanosomiasis and restenosis, and also include the design of derivatives to protect against rapid oxidation in vivo. Therefore, the large-scale production of cordycepin is currently desirable. As for the production method of cordycepin, the surface liquid culture using Cordyceps militaris mutant (G81-3) has been proven to be favorable for the higher production. However, the cordycepin productivity of the liquid surface culture was not so high because the culture period was long.
In this study, the cordycepin production by C. militaris mutant (G81-3) was examined using rotating disk contactor (RDC) for further improvement of the cordycepin productivity. When the specific biofilm surface area (total biofilm area divided by medium volume) of RDC was set at 5.1, the maximum cordycepin concentration obtained by RDC culture was 7.4 g/L, which was comparable with that of the liquid surface culture. Besides the cordycepin production rate was 2.7 g/(L*d), which was around 9 times that of the liquid surface culture. The RDC can be expected to be used as cordycepin production equipment because such excellent productivity was obtained.

Introduction
Utilization of renewable feedstocks for the production of bio-based chemicals such as D-lactic acid by engineering metabolic pathways in the yeast Saccharomyces cerevisiae has recently become an attractive option. In a previous study, we constructed efficient D-lactic acid producing S. cerevisiae YPH499/dPdA3-34/DLDH/1-18 by global metabolic engineering strategy1). However, the growth of the strain was strongly hampered by the presence of high concentrations of D-lactic acid, and thus it was not possible to achieve high titer D-lactic acid production without neutralization. In this study, expression of 11 endogenous transcription factors was optimized using the cocktail δ-integration strategy2) to improve D-lactic acid tolerance in YPH499/dPdA3-34/DLDH/1-18.
Results
DNA fragments, which contained 15 types of promoters and 11 types of transcription factor genes (HAA1, HSF1, MSN2, MSN4, PDR1, PDR3, RIM101, SFP1, SKN7, WAR1, and YAP1), were transformed into YPH499/dPdA3-34/DLDH/1-18. Among the resultant recombinant strains, YPH499/dPdA/DLDH/Lib-Tras143 with high lactic acid tolerance was selected. In non-neutralized flask fermentation, YPH499/dPdA/DLDH/Lib-Tras143 showed 1.2 fold higher D-lactic acid titer than YPH499/dPdA3-34/DLDH/1-18. In the transcriptional analysis by real-time PCR, the expression of 3 transcription factors HAA1, PDR3, and SFP1 was upregulated in YPH499/dPdA/DLDH/Lib-Tras143. Among them, the expression level of PDR3 was the highest, which may be a promising target for improvement of D-lactic acid tolerance in yeast S. cerevisiae.
Conclusion
The D-lactic acid tolerant strain YPH499/dPdA/DLDH/Lib-Tras143 has been successfully constructed by optimizing the expression of transcription factors. Transcriptional analysis revealed that 3 transcription factors (HAA1, PDR3 and SFP1) could function to improve lactic acid tolerance.
Acknowledgements
This work was partly supported by JSPS KAKENHI (grant number JP18K14069 and JP18KK0413).
Reference
1) R. Yamada et al.; Biotechnol Bioeng114:2075–2084 (2017)
2) R. Yamada et al.; Microb Cell Fact 9:32 (2010)
Lignocellulosic biomass is a promising feedstock for biofuel and other biochemical production to improve sustainability and the bioeconomy, and consists of cellulose and hemicellulose encapsulated in a lignin matrix. To convert biofuels from the lignocellulosic biomass, saccharification processes including hydrolysis are required before yeast fermentation. However it is one of serious problems that various toxic compounds coproduced by the saccharification processes. In this study, estimates of growth and fermentation inhibition during bioethanol fermentation were made using component profiles of corn cobs and corn stover at different degrees of hydrolysis. The component profiles were acquired by non-targeted gas chromatography mass spectrometry and targeted high-performance liquid chromatography(1). Correlations between the comprehensive analysis results and yeast growth and ethanol production were modeled very accurately by partial-least-squares regression analysis(1) and deep-learning. Acetate, apocynin, butyrovanillone, furfural, furyl hydroxymethyl ketone, m-methoxyacetophenone, palmitic acid, syringaldehyde, and xylose, were compounds with very variable importance in projection values and had negative correlation coefficients in the model. In fact, methoxyacetophenone, apocynin, and syringaldehyde inhibited fermentation more than furfural in equivalent concentration. Furthermore, deep-learning accurately estimated the time courses of cell growth and fermentation.
References
1) Watanabe K, Tachibana S, Konishi M. (2019) Bioresouc. Technol. 281: 260-268
The history of science and technology (S&T) that has been managed by the human race showed have much effect on the life patterns of the including ways to develop the quality of life and culture. Almost 100 years the era of industrialization indicating the existence of the dynamics of change, where social relationships between the community and the development S&T more and more quickly change. Consumptive behavior especially in exploring and exploitation of non-renewable natural resources already been surpassed the boundary of the natural capacity to meet the needs of human life. So that in realizing sustainable development has happened the transformation of economic development based on industry headed toward the sustainable development based on information. The era of this information demonstrate the ability a region and its people in doing efficiency and using information to achieve welfare competition on the situation at the global cooperation. So that the dynamics of the developments based on the knowledge are more complex. The development of roadmap of inclusive industrial systems, on the basis of locality characteristics for glucomannan industrialization from the Ammorphophallus Muelleri Blume (Indonesia Konjac) has been studied.
Introduction
Patchoulol is a sesquiterpene alcohol and an important natural product for the chemical industry. Although patchoulol is a component of plant patchouli essential oil, extraction from it is not a method suitable for large-scale production. Furthermore, using plants as resources for the production of sesquiterpene includes slow growth and varying composition depending on the geographical position and climate conditions.
Yeast Saccharomyces cerevisiae is widely used for production of various chemicals including the patchoulol by metabolic engineering. However, a limiting number of gene expression could be modified by conventional metabolic engineering in previous reports. Recently, global metabolic engineering strategy (GMES), which enables simultaneous modulation of many gene expressions, has been developed1). In this study, we constructed a mevalonate pathway engineered S. cerevisiae by GMES to realize efficient patchoulol production.
Methods
First, DNA fragments, which contained fifteen types of promoters and patchoulol synthase gene, were transformed into yeast YPH499. The resultant patchoulol synthase expressing strain was named YPH499/PAT167. Next, DNA fragments, which contained fifteen types of promoters and eight types of mevalonate pathway genes, were transformed into YPH499/PAT167. The resultant mevalonate pathway modified strain was named YPH499/PAT167/MVA442.
Results
As shown in the Figure, YPH499/PAT167 produced 8.4 mg/L patchoulol in 48 h cultivation, whereas YPH499/PAT167/MVA442 produced 39.5 mg/L patchoulol. By using the GMES, patchoulol production in a 48 h cultivation was improved 4.7 times, and the patchoulol volumetric productivity by YPH499/PAT167/MVA442 reached 19.8 mg/L/d.
To conclude, efficient patchoulol producing S. cerevisiae was successfully constructed by using GMES. GMES could also be applied for generating metabolically engineered S. cerevisiae capable of producing various bio-based chemicals.
Acknowledgements
This work was partly supported by JSPS KAKENHI (grant number JP18K14069 and JP18KK0413).
Reference
1) R. Yamada et al.; ACS Synth. Biol., 6, 659-666 (2017).

Shikimate pathway is one of the most important metabolic pathways in bacteria for synthesizing various aromatic compounds including aromatic amino acid.
Previous our study, we designed the metabolic pathway of Escherichia coli to enhance shikimate pathway by accumulating the intracellular PEP concentration and to produce chorismate derivatives in high yields. In order to avoid consuming PEP when uptake of glucose, we have generated a recombinant strain with replacing the endogenous phosphotransferase system (PTS) with GalP/Glk system (GGS) (PTS- GGS+ strain). In order to prevent leakage of carbon flux to the TCA cycle, we also disrupted two genes encoding pyruvate kinase (pykA, pykF), from PTS- GGS+ strain.
In this study, we designed two 窶廴etabolic Pathway Units窶・ first, we further modified the metabolism in E. coli to be confined the use of the glucose to the production of a target compound, MA. This pathway is the 窶廴A Production Unit窶・using glucose as a substrate. Second, to regain the capacity of cell growth, we introduced an exogenous Dahms pathway that supplements the metabolites necessary for cell growth with xylose into the strain. This pathway is the 窶廚ell Growth Unit窶・using xylose as a substrate. We attempted to produce MA in high yield by combining these two unitized pathways.
INTRODUCTION: Intestinal microbiome has recently been shown to play an important role in immune modulation, health and diseases. To understand the roles, great efforts have been made to develop in vitro models of the gastrointestinal human–microbe interface. However, most of reported models are microfluidics-based devices. Here, we fabricated bacteria-encapsulated microbeads to fabricate a simple, robust and tunable in vitro intestine model.
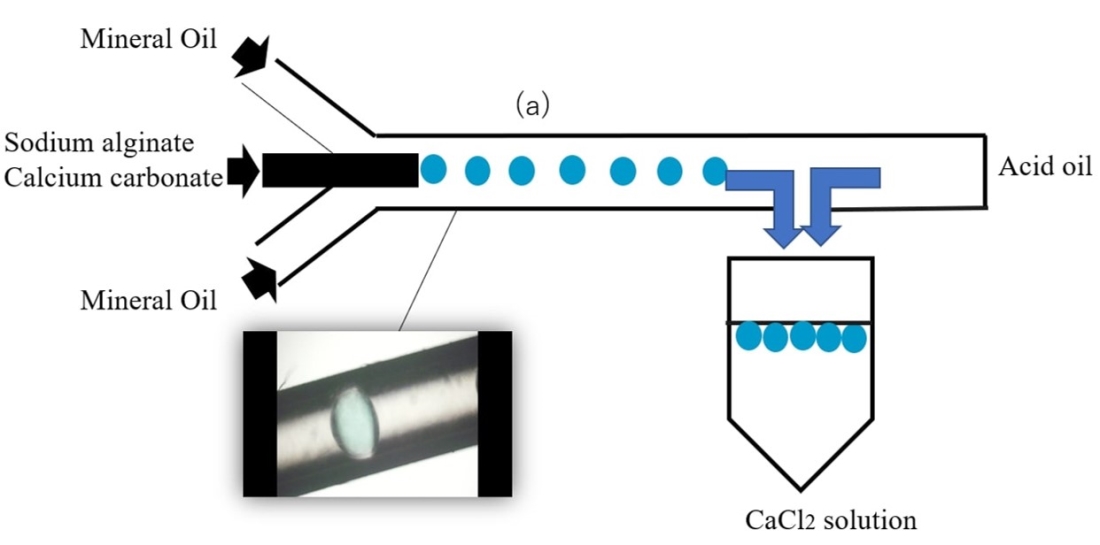
METHODS: A mixture solution of calcium carbonate, sodium alginate and E. coli was prepared and processed to form micro-droplets through a microfluidic water-in-oil emulsification. The microbeads were then gelated by changing pH(Fig.1). The surface of microbeads were sterilized, and then cultured in cell culture medium for 1 week. After 1 week, the presence or absence of leakage of E. coli, and the oxygen state inside the microbeads were characterized. Moreover, co-culture with a CACO-2 cell layer was performed to confirm the metabolism occurring in the presence of E. coli sealed in microbeads.
RESULTS & DISCUSSION : E. coli microbeads of different diameters were successfully fabricated by modulating the flow rate in different channels in the device. After 1 week of culture, no leakage of E. coli was observed, and it was confirmed that the E. coli in the microbeads were alive. In addition, the oxygen concentration in the microbeads was extremely reduced after 1 week, due to the anaerobic condition caused by E. coli oxygen consumption. The microbeads were immersed in tryptophan solution, and the production of indole was confirmed. CACO-2 cells are known to perform a sulfate conjugation reaction. Indoxyl sulfate produced from indole could be detected by co-culturing the E. coli microbeads with CACO-2 cells, further confirming the interaction between the CACO-2 cells with the E. coli encapsulated in the microbeads.
Twelve microorganisms were initially screened for their abilities to catalyze the biotransformation of phloretin. Two main products were identified in LC analysis after 1 hr of whole-cell bioconversion using Streptomyces avermitilis. To analyze the structural change of the product, TMS derivatization using BSTFA was done. After GC/MS analysis, product 1 is found to be hydroxylated in the A-ring of the phloretin with mass increase of 88 (192 to 280) in the A-ring fragment, and also for the whole phloretin molecule (562 to 650). Product 2 is also interpreted to be hydroxylated in the A-ring of the phloretin. But, its fragmentation pattern is different with product 2 with mass increase of 88 (179 to 267) in different A-ring fragment. Maximum conversion was 6.7 %, which was achieved for 1 hours of reaction, and the substrate (phloretin) and reaction product was completely metabolized after 3 hours of whole-cell reaction. Three kinds of Cytochrome P450 inhibitor, Coumarin, Erythromycin and Quinidine was added with 0.5mM final concentration, but, did not prevent the accumulation of the products, which means the reaction proceed not by P450 monoxygenase, possibly by other forms of oxygenase.
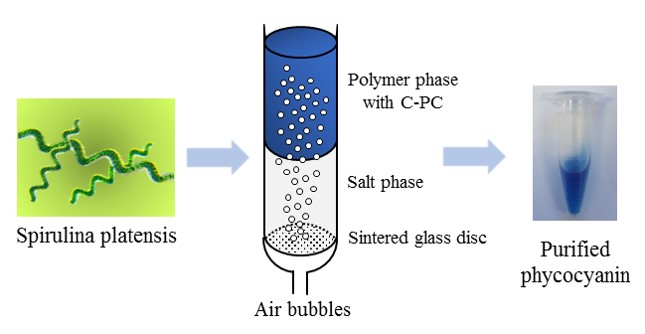
The purification and C-phycocyanin from Spirulina platensis was performed by employing the liquid biphasic flotation technique (LBF). LBF is a combination of both the liquid biphasic system (LBS) and solvent sublation, where the partitioning of the biomolecules in the system is enhanced through the effect of bubbling. The influence of various process parameters such as molecular weight of phase forming polymer, phase forming salt, phase composition, system pH, phase volume ratio, air flotation time and crude extract concentration were studied to optimize the C-phycocyanin recovery yield and purity. Desirable conditions for the purification of C-phycocyanin are found in the polymer-salt system consisting of polyethylene glycol (PEG) 4000 and potassium phosphate combination, where the increase in purification fold from 2.43 to 3.49 can be achieved in the optimized LBF. This technique has effectively enhanced the purification of C-phycocyanin in a cost effective and simple processing.